Code example: Wiener filter#
Introduction#
IFT starting point:
Typically, is a continuous field,
a discrete data vector. Particularly,
is not invertible.
IFT aims at inverting the above uninvertible problem in the best possible way using Bayesian statistics.
NIFTy (Numerical Information Field Theory) is a Python framework in which IFT problems can be tackled easily.
Main Interfaces:
Spaces: Cartesian, 2-Spheres (Healpix, Gauss-Legendre) and their respective harmonic spaces.
Fields: Defined on spaces.
Operators: Acting on fields.
Wiener filter on one-dimensional fields#
Assumptions#
,
linear operator.
,
where
are positive definite matrices.
Posterior#
The Posterior is given by:
where
$$
Let us implement this in NIFTy!
In NIFTy#
We assume statistical homogeneity and isotropy. Therefore the signal covariance
is diagonal in harmonic space, and is described by a one-dimensional power spectrum, assumed here as $
P_0 = 0.2, k_0 = 5, \gamma = 4$.
.
Number of data points
.
reconstruction in harmonic space.
Response operator: $
$
N_pixels = 512 # Number of pixels
def pow_spec(k):
P0, k0, gamma = [.2, 5, 4]
return P0 / ((1. + (k/k0)**2)**(gamma / 2))
Implementation#
Import Modules#
%matplotlib inline
import numpy as np
import nifty8 as ift
import matplotlib.pyplot as plt
plt.rcParams['figure.dpi'] = 100
Implement Propagator#
def Curvature(R, N, Sh):
IC = ift.GradientNormController(iteration_limit=50000,
tol_abs_gradnorm=0.1)
# WienerFilterCurvature is (R.adjoint*N.inverse*R + Sh.inverse) plus some handy
# helper methods.
return ift.WienerFilterCurvature(R,N,Sh,iteration_controller=IC,iteration_controller_sampling=IC)
Conjugate Gradient Preconditioning#
is defined via: $
D
j
D^{-1}$. This is done numerically (algorithm: Conjugate Gradient).
Generate Mock data#
Generate a field
and
with given covariances.
Calculate
.
s_space = ift.RGSpace(N_pixels)
h_space = s_space.get_default_codomain()
HT = ift.HarmonicTransformOperator(h_space, target=s_space)
# Operators
Sh = ift.create_power_operator(h_space, power_spectrum=pow_spec, sampling_dtype=float)
R = HT # @ ift.create_harmonic_smoothing_operator((h_space,), 0, 0.02)
# Fields and data
sh = Sh.draw_sample()
noiseless_data=R(sh)
noise_amplitude = np.sqrt(0.2)
N = ift.ScalingOperator(s_space, noise_amplitude**2, float)
n = ift.Field.from_random(domain=s_space, random_type='normal',
std=noise_amplitude, mean=0)
d = noiseless_data + n
j = R.adjoint_times(N.inverse_times(d))
curv = Curvature(R=R, N=N, Sh=Sh)
D = curv.inverse
Run Wiener Filter#
m = D(j)
Results#
# Get signal data and reconstruction data
s_data = HT(sh).val
m_data = HT(m).val
d_data = d.val
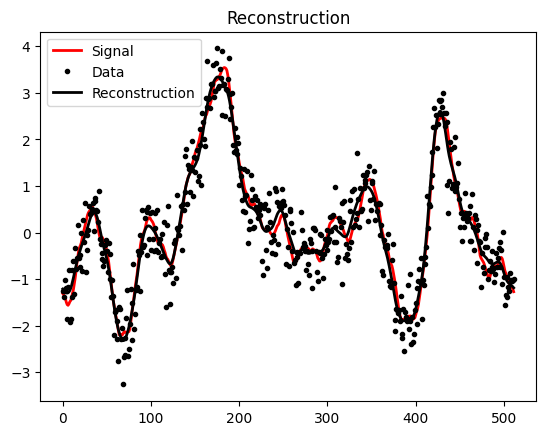
plt.plot(s_data, 'r', label="Signal", linewidth=2)
plt.plot(d_data, 'k.', label="Data")
plt.plot(m_data, 'k', label="Reconstruction",linewidth=2)
plt.title("Reconstruction")
plt.legend()
plt.show()
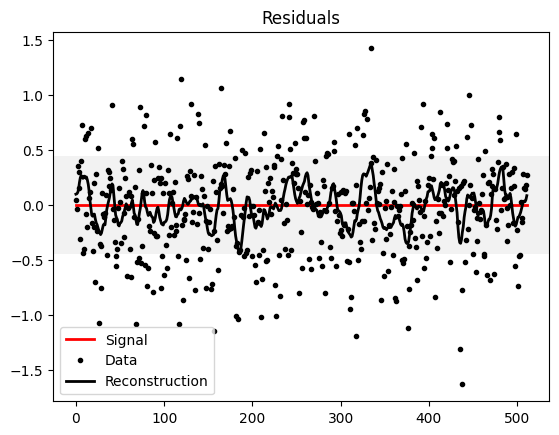
plt.plot(s_data - s_data, 'r', label="Signal", linewidth=2)
plt.plot(d_data - s_data, 'k.', label="Data")
plt.plot(m_data - s_data, 'k', label="Reconstruction",linewidth=2)
plt.axhspan(-noise_amplitude,noise_amplitude, facecolor='0.9', alpha=.5)
plt.title("Residuals")
plt.legend()
plt.show()
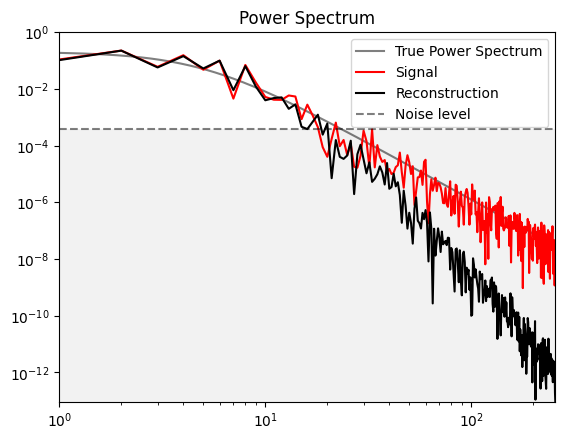
Power Spectrum#
s_power_data = ift.power_analyze(sh).val
m_power_data = ift.power_analyze(m).val
plt.loglog()
plt.xlim(1, int(N_pixels/2))
ymin = min(m_power_data)
plt.ylim(ymin, 1)
xs = np.arange(1,int(N_pixels/2),.1)
plt.plot(xs, pow_spec(xs), label="True Power Spectrum", color='k',alpha=0.5)
plt.plot(s_power_data, 'r', label="Signal")
plt.plot(m_power_data, 'k', label="Reconstruction")
plt.axhline(noise_amplitude**2 / N_pixels, color="k", linestyle='--', label="Noise level", alpha=.5)
plt.axhspan(noise_amplitude**2 / N_pixels, ymin, facecolor='0.9', alpha=.5)
plt.title("Power Spectrum")
plt.legend()
plt.show()
Wiener Filter on Incomplete Data#
# Operators
Sh = ift.create_power_operator(h_space, power_spectrum=pow_spec, sampling_dtype=float)
N = ift.ScalingOperator(s_space, noise_amplitude**2, sampling_dtype=float)
# R is defined below
# Fields
sh = Sh.draw_sample()
s = HT(sh)
n = ift.Field.from_random(domain=s_space, random_type='normal',
std=noise_amplitude, mean=0)
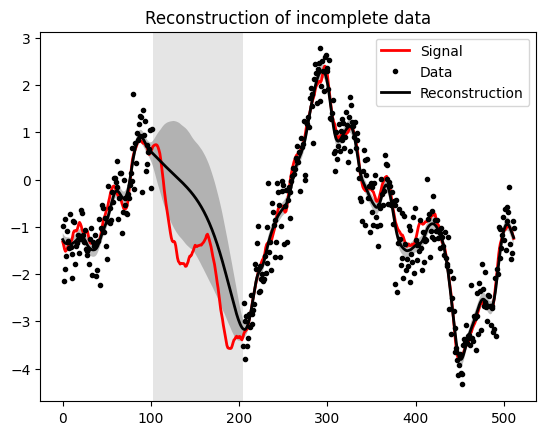
Partially Lose Data#
l = int(N_pixels * 0.2)
h = int(N_pixels * 0.2 * 2)
mask = np.full(s_space.shape, 1.)
mask[l:h] = 0
mask = ift.Field.from_raw(s_space, mask)
R = ift.DiagonalOperator(mask) @ HT
n = n.val_rw()
n[l:h] = 0
n = ift.Field.from_raw(s_space, n)
d = R(sh) + n
curv = Curvature(R=R, N=N, Sh=Sh)
D = curv.inverse
j = R.adjoint_times(N.inverse_times(d))
m = D(j)
Compute Uncertainty#
m_mean, m_var = ift.probe_with_posterior_samples(curv, HT, 200, np.float64)
Get data#
# Get signal data and reconstruction data
s_data = s.val
m_data = HT(m).val
m_var_data = m_var.val
uncertainty = np.sqrt(m_var_data)
d_data = d.val_rw()
# Set lost data to NaN for proper plotting
d_data[d_data == 0] = np.nan
plt.axvspan(l, h, facecolor='0.8',alpha=0.5)
plt.fill_between(range(N_pixels), m_data - uncertainty, m_data + uncertainty, facecolor='0.5', alpha=0.5)
plt.plot(s_data, 'r', label="Signal", alpha=1, linewidth=2)
plt.plot(d_data, 'k.', label="Data")
plt.plot(m_data, 'k', label="Reconstruction", linewidth=2)
plt.title("Reconstruction of incomplete data")
plt.legend()
<matplotlib.legend.Legend at 0x7ff34868b350>
Wiener filter on two-dimensional field#
N_pixels = 256 # Number of pixels
sigma2 = 2. # Noise variance
def pow_spec(k):
P0, k0, gamma = [.2, 2, 4]
return P0 * (1. + (k/k0)**2)**(-gamma/2)
s_space = ift.RGSpace([N_pixels, N_pixels])
h_space = s_space.get_default_codomain()
HT = ift.HarmonicTransformOperator(h_space,s_space)
# Operators
Sh = ift.create_power_operator(h_space, power_spectrum=pow_spec, sampling_dtype=float)
N = ift.ScalingOperator(s_space, sigma2, sampling_dtype=float)
# Fields and data
sh = Sh.draw_sample()
n = ift.Field.from_random(domain=s_space, random_type='normal',
std=np.sqrt(sigma2), mean=0)
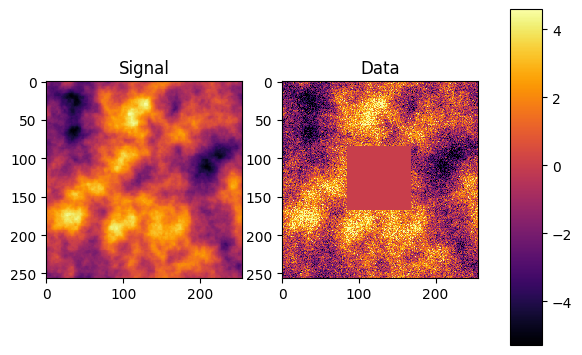
# Lose some data
l = int(N_pixels * 0.33)
h = int(N_pixels * 0.33 * 2)
mask = np.full(s_space.shape, 1.)
mask[l:h,l:h] = 0.
mask = ift.Field.from_raw(s_space, mask)
R = ift.DiagonalOperator(mask)(HT)
n = n.val_rw()
n[l:h, l:h] = 0
n = ift.Field.from_raw(s_space, n)
curv = Curvature(R=R, N=N, Sh=Sh)
D = curv.inverse
d = R(sh) + n
j = R.adjoint_times(N.inverse_times(d))
# Run Wiener filter
m = D(j)
# Uncertainty
m_mean, m_var = ift.probe_with_posterior_samples(curv, HT, 20, np.float64)
# Get data
s_data = HT(sh).val
m_data = HT(m).val
m_var_data = m_var.val
d_data = d.val
uncertainty = np.sqrt(np.abs(m_var_data))
cmap = ['magma', 'inferno', 'plasma', 'viridis'][1]
mi = np.min(s_data)
ma = np.max(s_data)
fig, axes = plt.subplots(1, 2)
data = [s_data, d_data]
caption = ["Signal", "Data"]
for ax in axes.flat:
im = ax.imshow(data.pop(0), interpolation='nearest', cmap=cmap, vmin=mi,
vmax=ma)
ax.set_title(caption.pop(0))
fig.subplots_adjust(right=0.8)
cbar_ax = fig.add_axes([0.85, 0.15, 0.05, 0.7])
fig.colorbar(im, cax=cbar_ax)
<matplotlib.colorbar.Colorbar at 0x7ff348a13ed0>
mi = np.min(s_data)
ma = np.max(s_data)
fig, axes = plt.subplots(3, 2, figsize=(10, 15))
sample = HT(curv.draw_sample(from_inverse=True)+m).val
post_mean = (m_mean + HT(m)).val
data = [s_data, m_data, post_mean, sample, s_data - m_data, uncertainty]
caption = ["Signal", "Reconstruction", "Posterior mean", "Sample", "Residuals", "Uncertainty Map"]
for ax in axes.flat:
im = ax.imshow(data.pop(0), interpolation='nearest', cmap=cmap, vmin=mi, vmax=ma)
ax.set_title(caption.pop(0))
fig.subplots_adjust(right=0.8)
cbar_ax = fig.add_axes([.85, 0.15, 0.05, 0.7])
fig.colorbar(im, cax=cbar_ax)
<matplotlib.colorbar.Colorbar at 0x7ff344a0c610>
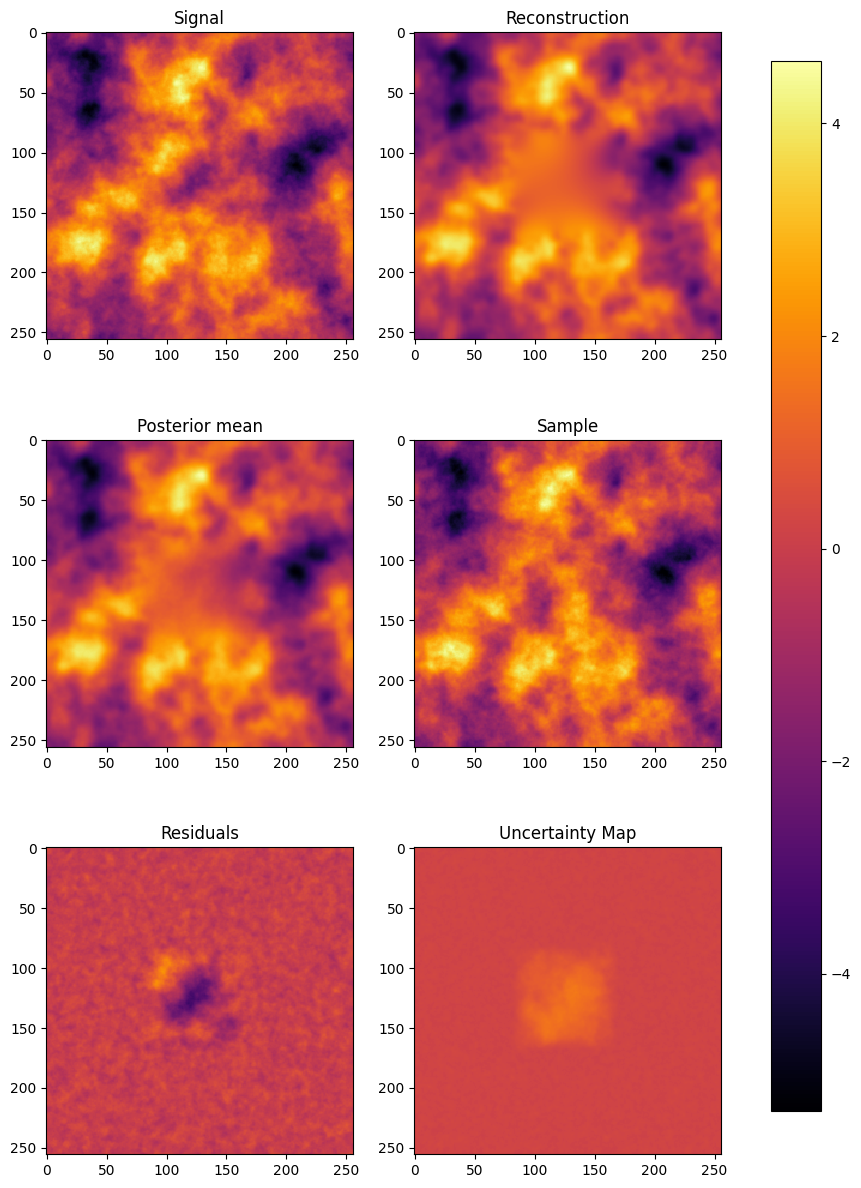
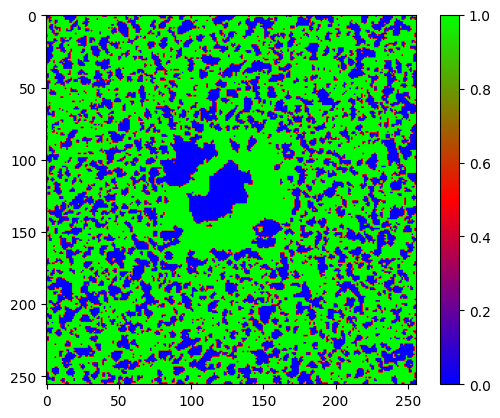
Is the uncertainty map reliable?#
precise = (np.abs(s_data-m_data) < uncertainty)
print("Error within uncertainty map bounds: " + str(np.sum(precise) * 100 / N_pixels**2) + "%")
plt.imshow(precise.astype(float), cmap="brg")
plt.colorbar()
Error within uncertainty map bounds: 67.71392822265625%
<matplotlib.colorbar.Colorbar at 0x7ff34421f250>