Showcasing the Correlated Field model#
Skip to Parameter Showcases for the meat/veggies ;)
The field model roughly works like this:
f = HT( A * zero_mode * xi ) + offset
A is a spectral power field which is constructed from power spectra that hold on subdomains of the target domain.
It is scaled by a zero mode operator and then pointwise multiplied by a gaussian excitation field, yielding
a representation of the field in harmonic space.
It is then transformed into the target real space and a offset added.
The power spectra A is constructed of are in turn constructed as the sum of a power law component
and an integrated Wiener process whose amplitude and roughness can be set.
Preliminaries#
Setup code#
%matplotlib inline
import nifty8 as ift
import matplotlib.pyplot as plt
import numpy as np
plt.rcParams['figure.dpi'] = 100
n_pix = 256
x_space = ift.RGSpace(n_pix)
ift.random.push_sseq_from_seed(1)
# Plotting routine
def plot(fields, spectra, title=None):
# Plotting preparation is normally handled by nifty8.Plot
# It is done manually here to be able to tweak details
# Fields are assumed to have identical domains
fig = plt.figure(tight_layout=True, figsize=(10, 3))
if title is not None:
fig.suptitle(title, fontsize=14)
# Field
ax1 = fig.add_subplot(1, 2, 1)
ax1.axhline(y=0., color='k', linestyle='--', alpha=0.25)
for field in fields:
dom = field.domain[0]
xcoord = np.arange(dom.shape[0]) * dom.distances[0]
ax1.plot(xcoord, field.val)
ax1.set_xlim(xcoord[0], xcoord[-1])
ax1.set_ylim(-5., 5.)
ax1.set_xlabel('x')
ax1.set_ylabel('f(x)')
ax1.set_title('Field realizations')
# Spectrum
ax2 = fig.add_subplot(1, 2, 2)
for spectrum in spectra:
xcoord = spectrum.domain[0].k_lengths
ycoord = spectrum.val_rw()
ycoord[0] = ycoord[1]
ax2.plot(xcoord, ycoord)
ax2.set_ylim(1e-6, 10.)
ax2.set_xscale('log')
ax2.set_yscale('log')
ax2.set_xlabel('k')
ax2.set_ylabel('p(k)')
ax2.set_title('Power Spectrum')
fig.align_labels()
plt.show()
# Helper: draw main sample
main_sample = None
def init_model(m_pars, fl_pars, matern=False):
global main_sample
cf = ift.CorrelatedFieldMaker(m_pars["prefix"])
cf.set_amplitude_total_offset(m_pars["offset_mean"], m_pars["offset_std"])
cf.add_fluctuations_matern(**fl_pars) if matern else cf.add_fluctuations(**fl_pars)
field = cf.finalize(prior_info=0)
main_sample = ift.from_random(field.domain)
print("model domain keys:", field.domain.keys())
# Helper: field and spectrum from parameter dictionaries + plotting
def eval_model(m_pars, fl_pars, title=None, samples=None, matern=False):
cf = ift.CorrelatedFieldMaker(m_pars["prefix"])
cf.set_amplitude_total_offset(m_pars["offset_mean"], m_pars["offset_std"])
cf.add_fluctuations_matern(**fl_pars) if matern else cf.add_fluctuations(**fl_pars)
field = cf.finalize(prior_info=0)
spectrum = cf.amplitude
if samples is None:
samples = [main_sample]
field_realizations = [field(s) for s in samples]
spectrum_realizations = [spectrum.force(s) for s in samples]
plot(field_realizations, spectrum_realizations, title)
def gen_samples(key_to_vary):
if key_to_vary is None:
return [main_sample]
dct = main_sample.to_dict()
subdom_to_vary = dct.pop(key_to_vary).domain
samples = []
for i in range(8):
d = dct.copy()
d[key_to_vary] = ift.from_random(subdom_to_vary)
samples.append(ift.MultiField.from_dict(d))
return samples
def vary_parameter(parameter_key, values, samples_vary_in=None, matern=False):
s = gen_samples(samples_vary_in)
for v in values:
if parameter_key in cf_make_pars.keys():
m_pars = {**cf_make_pars, parameter_key: v}
eval_model(m_pars, cf_x_fluct_pars, f"{parameter_key} = {v}", s, matern)
else:
fl_pars = {**cf_x_fluct_pars, parameter_key: v}
eval_model(cf_make_pars, fl_pars, f"{parameter_key} = {v}", s, matern)
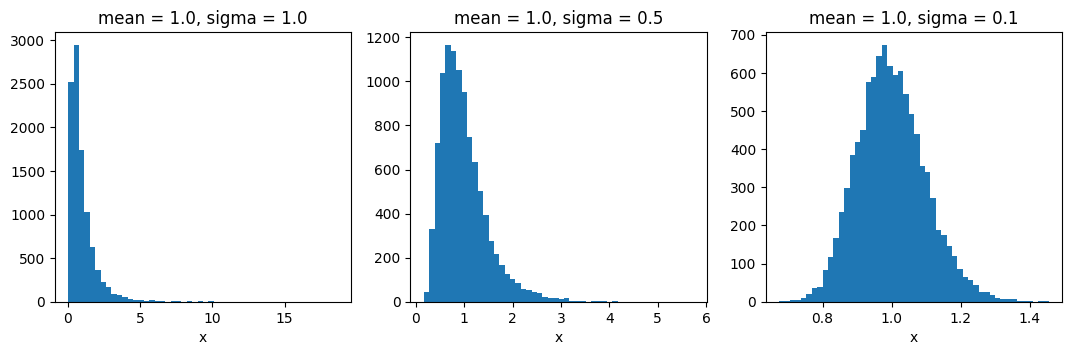
Before the Action: The Moment-Matched Log-Normal Distribution#
Many properties of the correlated field are modelled as being lognormally distributed.
The distribution models are parametrized via their means and standard-deviations (first and second position in tuple).
To get a feeling of how the ratio of the mean and stddev parameters influences the distribution shape,
here are a few example histograms: (observe the x-axis!)
fig = plt.figure(figsize=(13, 3.5))
mean = 1.0
sigmas = [1.0, 0.5, 0.1]
for i in range(3):
op = ift.LognormalTransform(mean=mean, sigma=sigmas[i],
key='foo', N_copies=0)
op_samples = np.array(
[op(s).val for s in [ift.from_random(op.domain) for i in range(10000)]])
ax = fig.add_subplot(1, 3, i + 1)
ax.hist(op_samples, bins=50)
ax.set_title(f"mean = {mean}, sigma = {sigmas[i]}")
ax.set_xlabel('x')
del op_samples
plt.show()
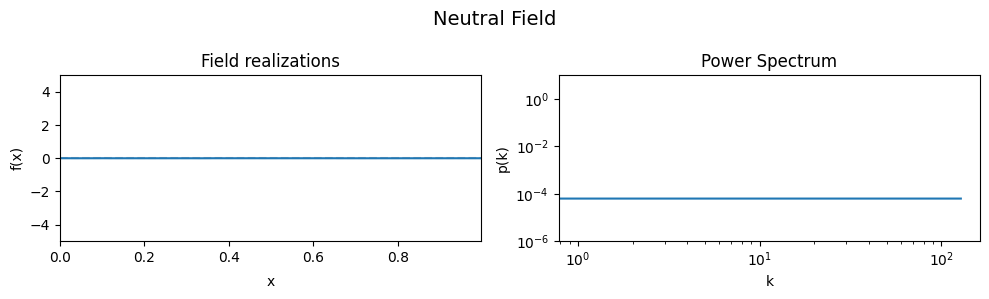
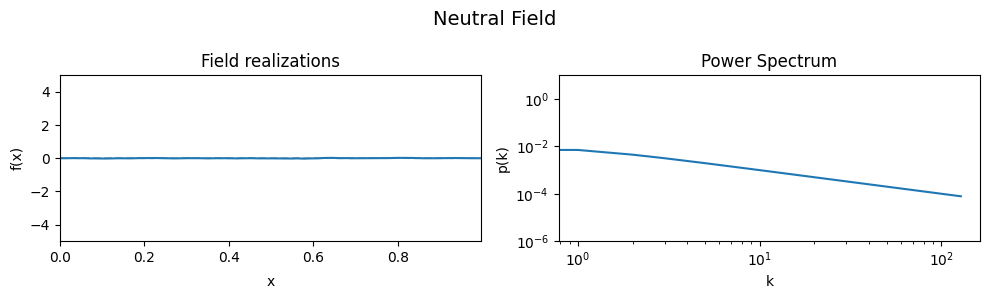
The Neutral Field#
To demonstrate the effect of all parameters, first a ‘neutral’ set of parameters is defined which then are varied one by one, showing the effect of the variation on the generated field realizations and the underlying power spectrum from which they were drawn.
As a neutral field, a model with a white power spectrum and vanishing spectral power was chosen.
# Neutral model parameters yielding a quasi-constant field
cf_make_pars = {
'offset_mean': 0.,
'offset_std': (1e-3, 1e-16),
'prefix': ''
}
cf_x_fluct_pars = {
'target_subdomain': x_space,
'fluctuations': (1e-3, 1e-16),
'flexibility': (1e-3, 1e-16),
'asperity': (1e-3, 1e-16),
'loglogavgslope': (0., 1e-16)
}
init_model(cf_make_pars, cf_x_fluct_pars)
model domain keys: ('asperity', 'flexibility', 'fluctuations', 'loglogavgslope', 'spectrum', 'xi', 'zeromode')
# Show neutral field
eval_model(cf_make_pars, cf_x_fluct_pars, "Neutral Field")
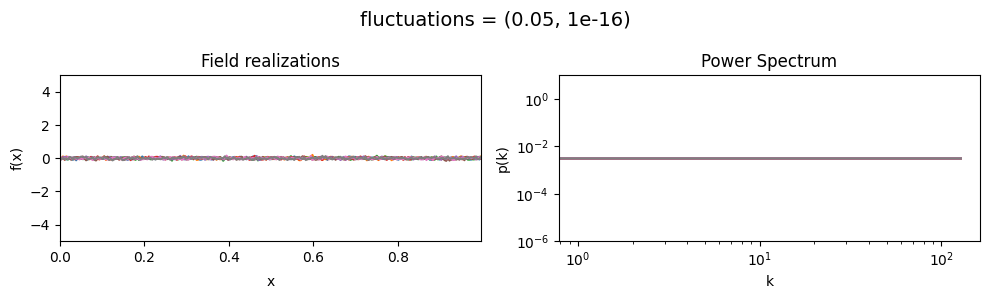
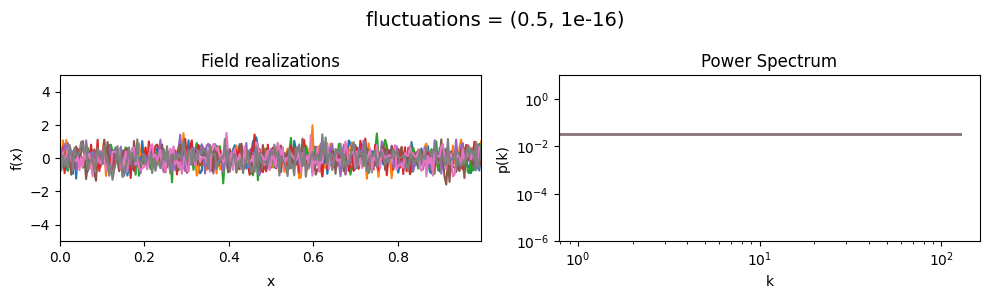
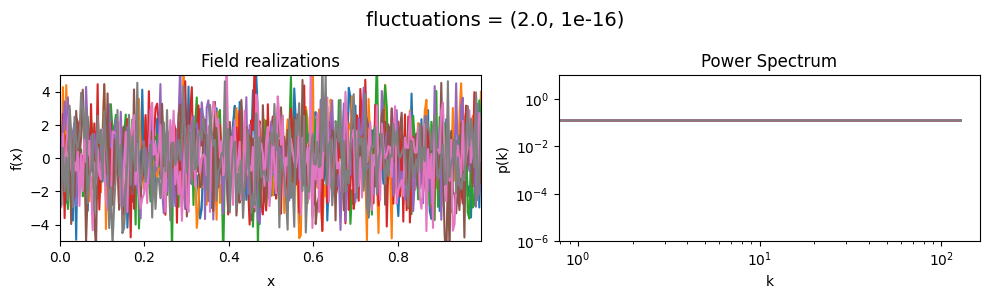
The fluctuations parameters of add_fluctuations()#
determine the amplitude of variations along the field dimension
for which add_fluctuations is called.
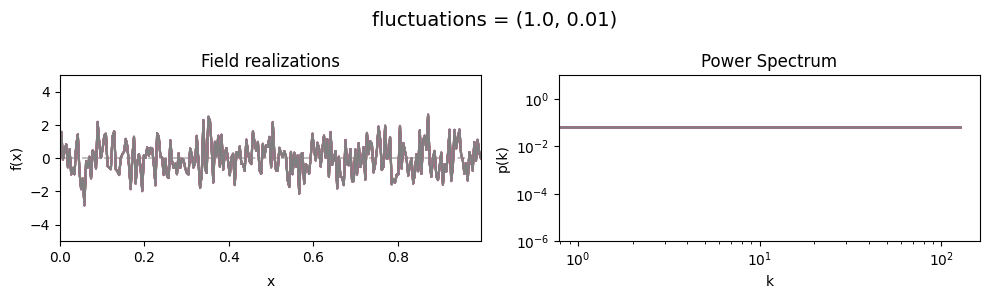
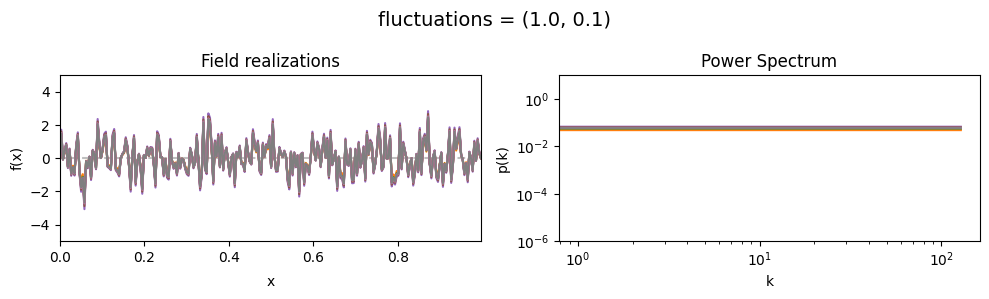
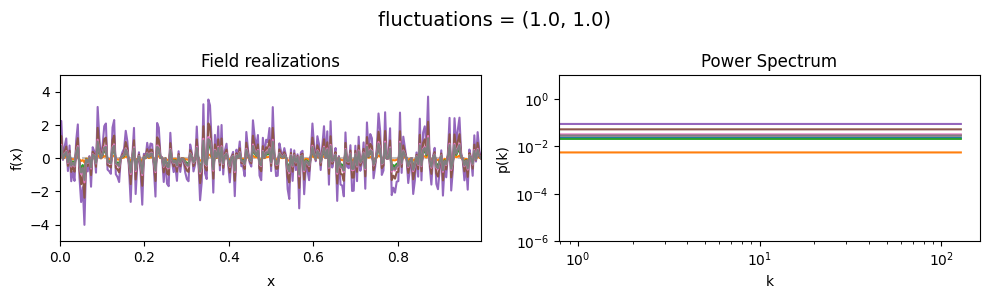
fluctuations[0] set the average amplitude of the fields fluctuations along the given dimension,
fluctuations[1] sets the width and shape of the amplitude distribution.
The amplitude is modelled as being log-normally distributed,
see The Moment-Matched Log-Normal Distribution above for details.
fluctuations mean:#
vary_parameter('fluctuations', [(0.05, 1e-16), (0.5, 1e-16), (2., 1e-16)], samples_vary_in='xi')
fluctuations std:#
vary_parameter('fluctuations', [(1., 0.01), (1., 0.1), (1., 1.)], samples_vary_in='fluctuations')
cf_x_fluct_pars['fluctuations'] = (1., 1e-16)
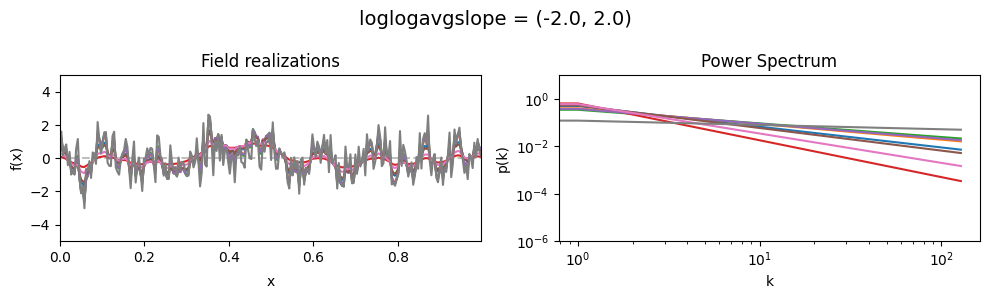
The loglogavgslope parameters of add_fluctuations()#
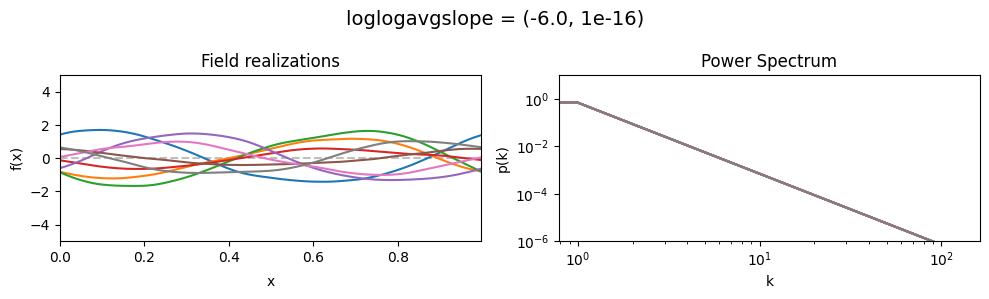
determine the slope of the loglog-linear (power law) component of the power spectrum.
The slope is modelled to be normally distributed.
loglogavgslope mean:#
vary_parameter('loglogavgslope', [(-6., 1e-16), (-2., 1e-16), (2., 1e-16)], samples_vary_in='xi')
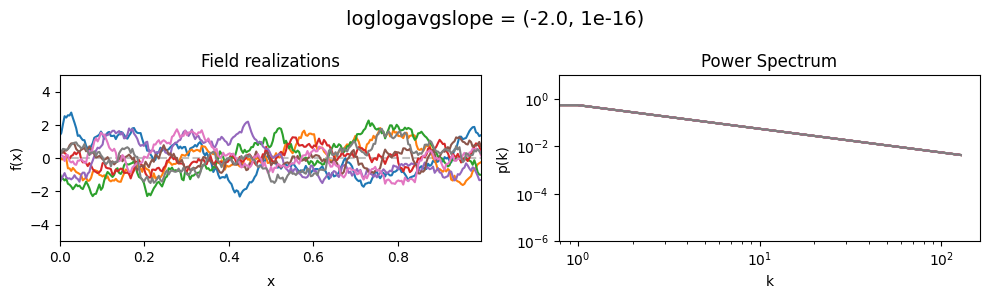
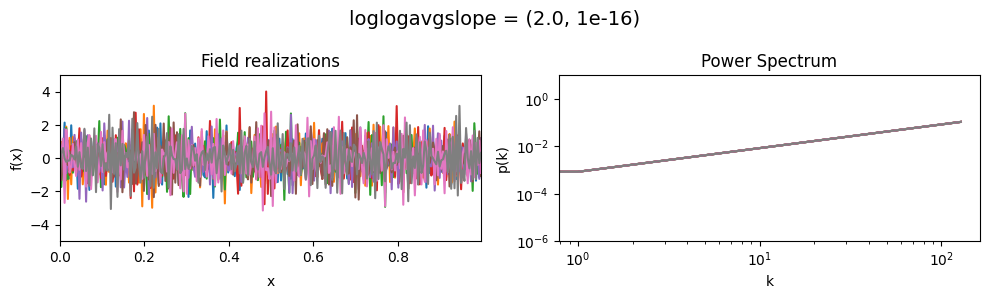
loglogavgslope std:#
vary_parameter('loglogavgslope', [(-2., 0.02), (-2., 0.2), (-2., 2.0)], samples_vary_in='loglogavgslope')
cf_x_fluct_pars['loglogavgslope'] = (-2., 1e-16)
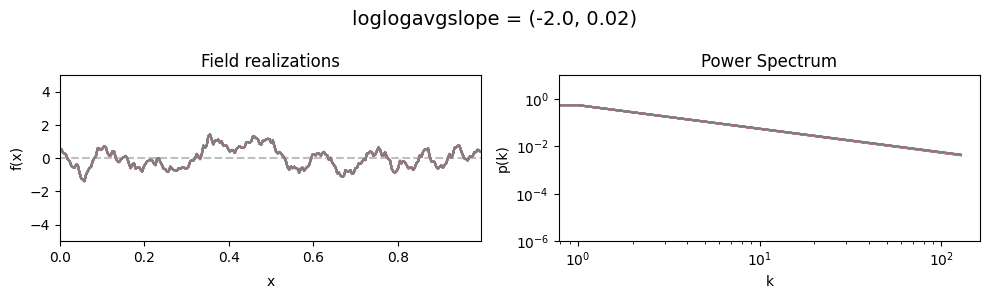
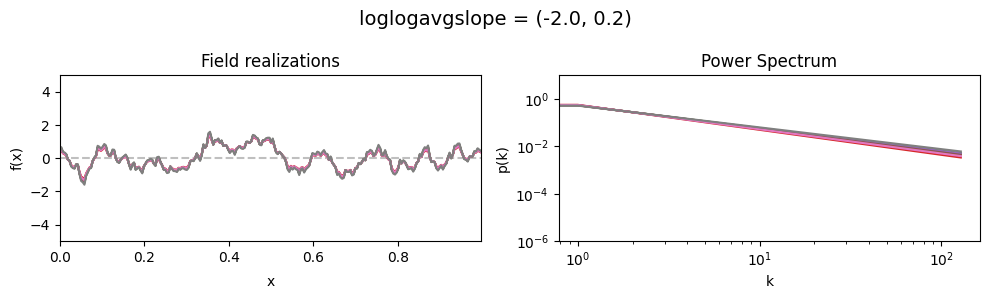
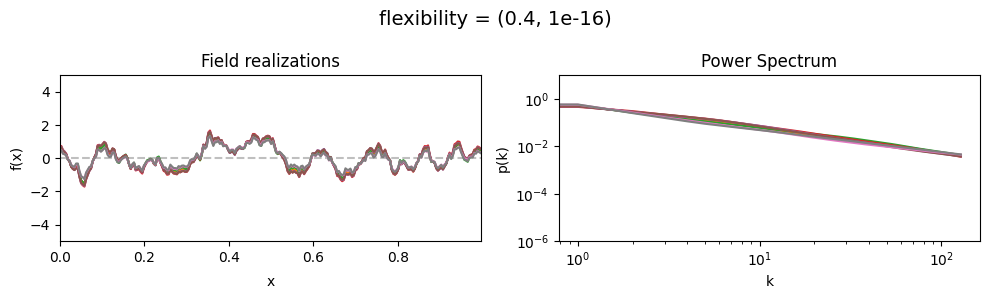
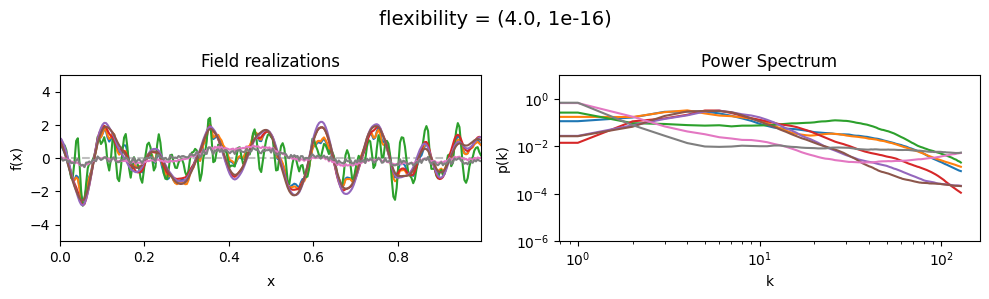
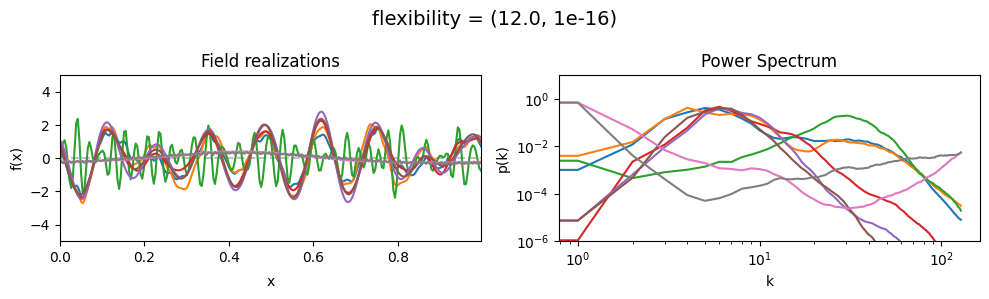
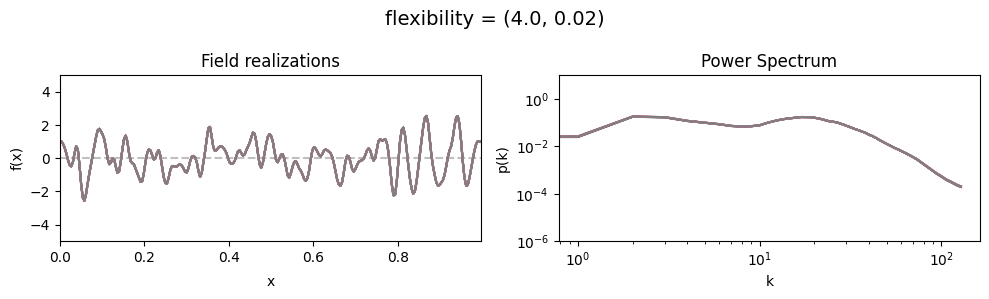
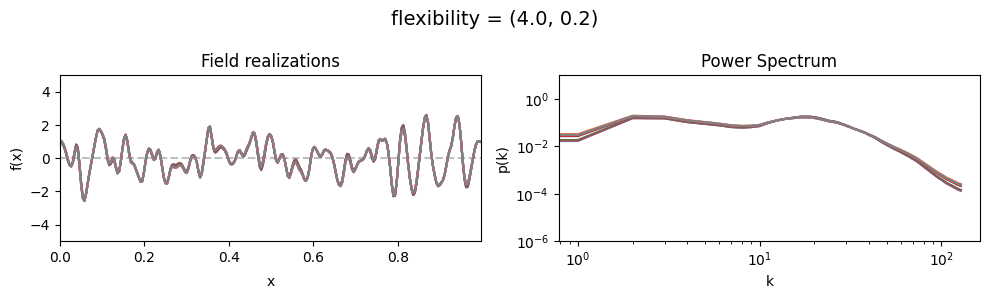
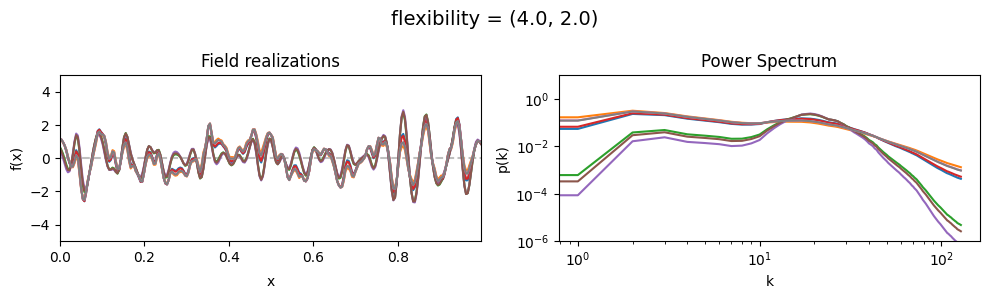
The flexibility parameters of add_fluctuations()#
determine the amplitude of the integrated Wiener process component of the power spectrum (how strong the power spectrum varies besides the power-law).
flexibility[0] sets the average amplitude of the i.g.p. component,
flexibility[1] sets how much the amplitude can vary.
These two parameters feed into a moment-matched log-normal distribution model,
see above for a demo of its behavior.
flexibility mean:#
vary_parameter('flexibility', [(0.4, 1e-16), (4.0, 1e-16), (12.0, 1e-16)], samples_vary_in='spectrum')
flexibility std:#
vary_parameter('flexibility', [(4., 0.02), (4., 0.2), (4., 2.)], samples_vary_in='flexibility')
cf_x_fluct_pars['flexibility'] = (4., 1e-16)
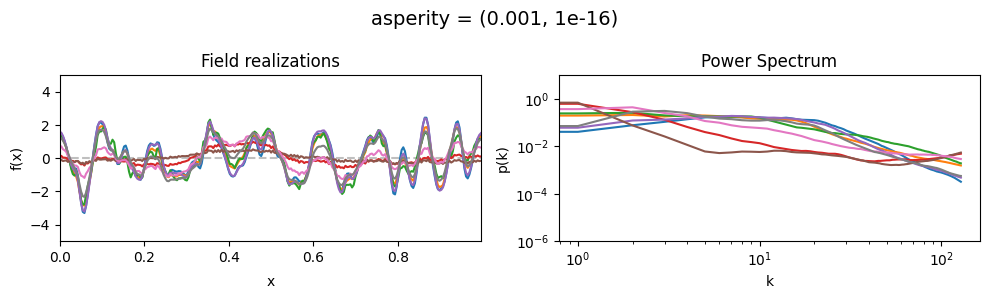
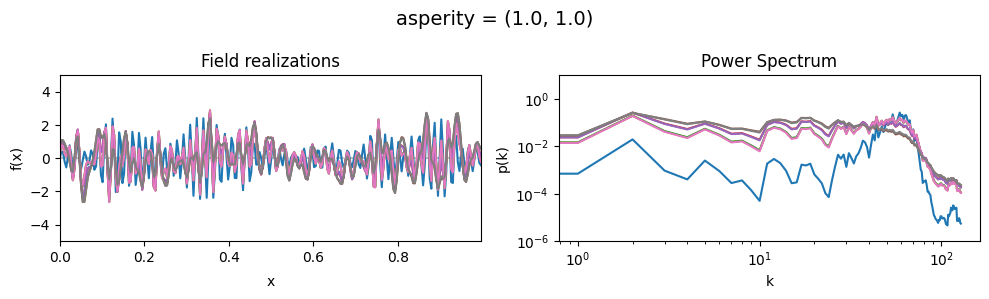
The asperity parameters of add_fluctuations()#
asperity determines how rough the integrated Wiener process component of the power spectrum is.
asperity[0] sets the average roughness, asperity[1] sets how much the roughness can vary.
These two parameters feed into a moment-matched log-normal distribution model,
see above for a demo of its behavior.
asperity mean:#
vary_parameter('asperity', [(0.001, 1e-16), (1.0, 1e-16), (5., 1e-16)], samples_vary_in='spectrum')
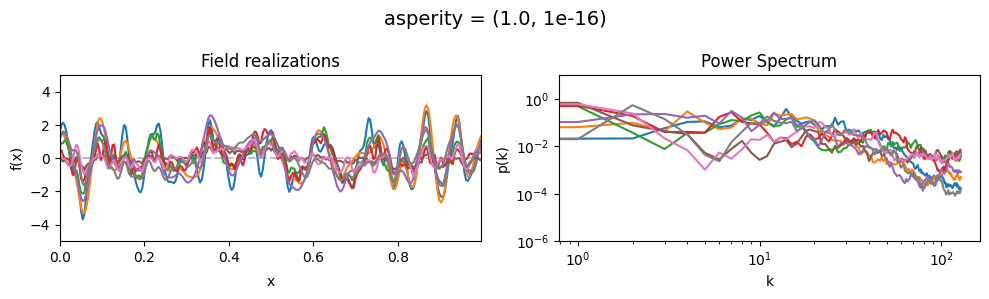
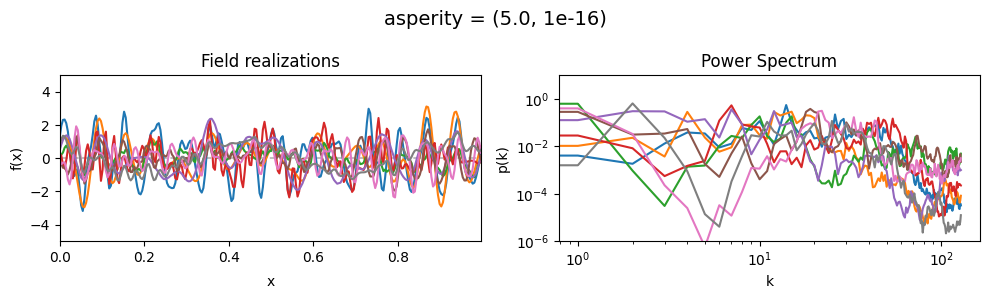
asperity std:#
vary_parameter('asperity', [(1., 0.01), (1., 0.1), (1., 1.)], samples_vary_in='asperity')
cf_x_fluct_pars['asperity'] = (1., 1e-16)
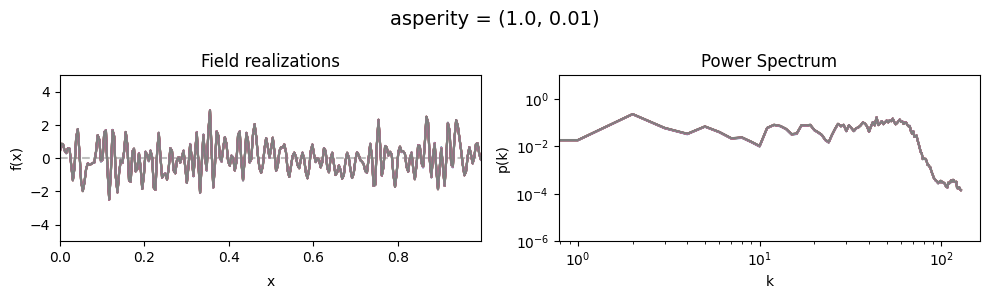
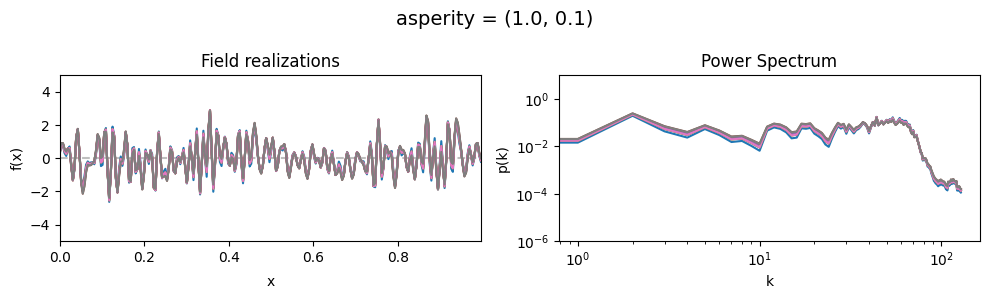
The offset_mean parameter of CorrelatedFieldMaker()#
The offset_mean parameter defines a global additive offset on the field realizations.
If the field is used for a lognormal model f = field.exp(), this acts as a global signal magnitude offset.
# Reset model to neutral
cf_x_fluct_pars['fluctuations'] = (1e-3, 1e-16)
cf_x_fluct_pars['flexibility'] = (1e-3, 1e-16)
cf_x_fluct_pars['asperity'] = (1e-3, 1e-16)
cf_x_fluct_pars['loglogavgslope'] = (1e-3, 1e-16)
vary_parameter('offset_mean', [3., 0., -2.])
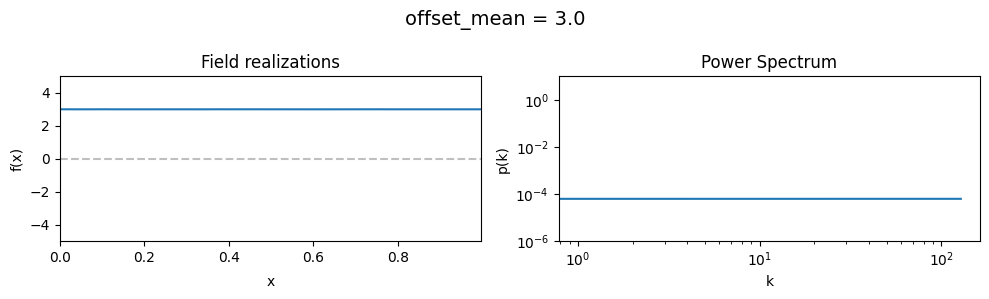
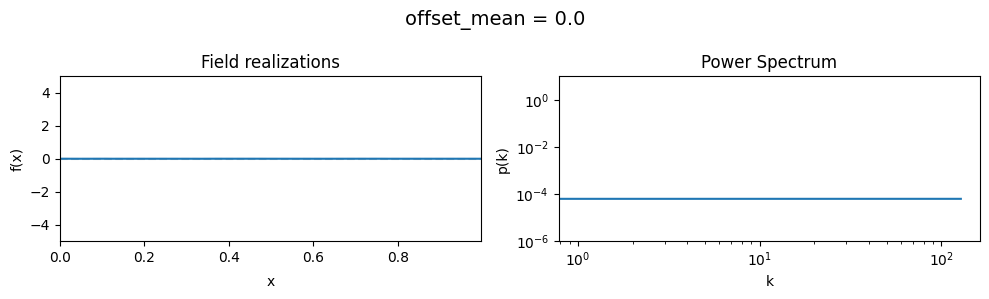
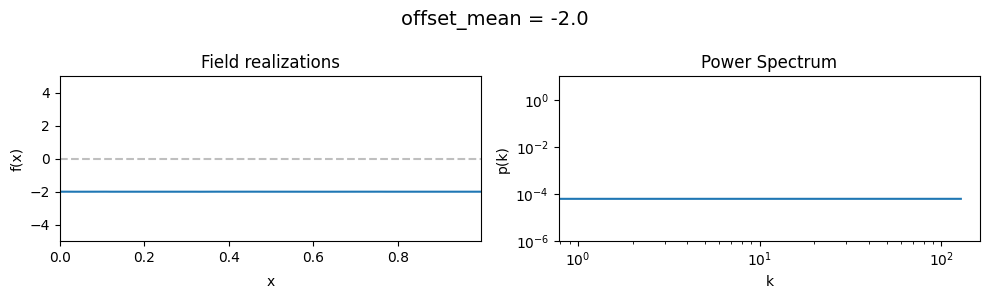
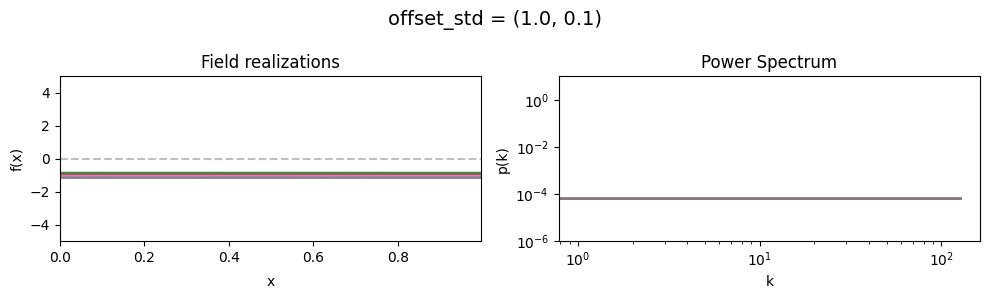
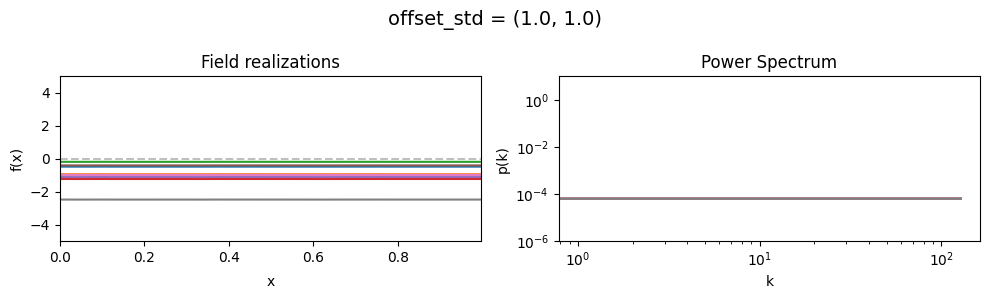
The offset_std parameters of CorrelatedFieldMaker()#
Variation of the global offset of the field are modelled as being log-normally distributed.
See The Moment-Matched Log-Normal Distribution above for details.
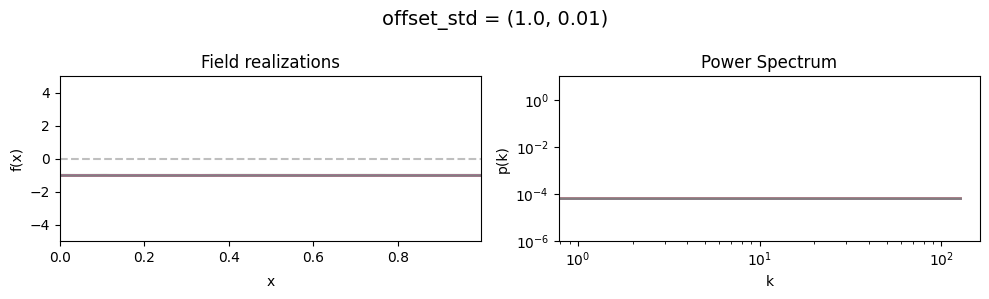
The offset_std[0] parameter sets how much NIFTy will vary the offset on average.
The offset_std[1] parameters defines the with and shape of the offset variation distribution.
offset_std mean:#
vary_parameter('offset_std', [(1e-16, 1e-16), (0.5, 1e-16), (2., 1e-16)], samples_vary_in='xi')
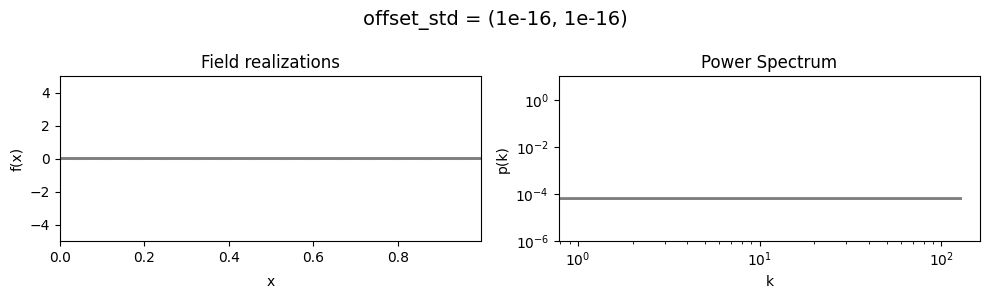
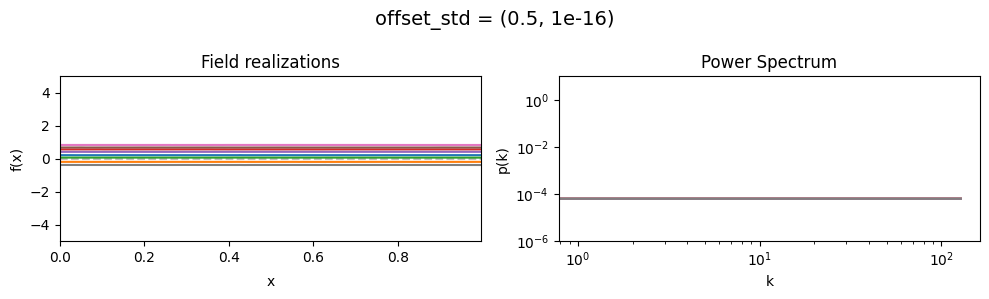
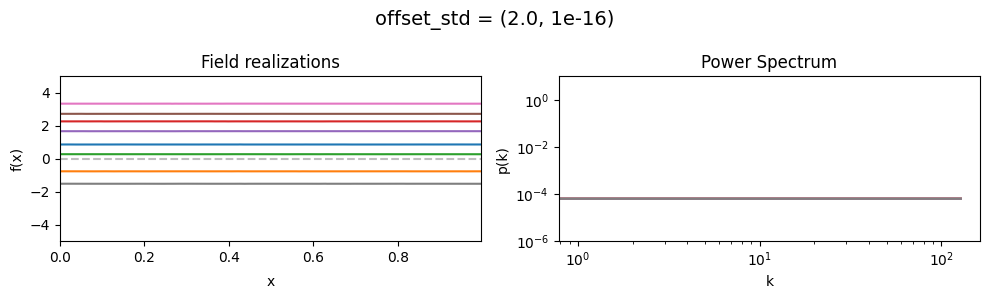
offset_std std:#
vary_parameter('offset_std', [(1., 0.01), (1., 0.1), (1., 1.)], samples_vary_in='zeromode')
Matern fluctuation kernels#
The correlated fields model also supports parametrizing the power spectra of field dimensions using Matern kernels. In the following, the effects of their parameters are demonstrated.
Contrary to the field fluctuations parametrization showed above, the Matern kernel
parameters show strong interactions. For example, the field amplitude does not only depend on the
amplitude scaling parameter scale, but on the combination of all three parameters scale,
cutoff and loglogslope.
# Neutral model parameters yielding a quasi-constant field
cf_make_pars = {
'offset_mean': 0.,
'offset_std': (1e-3, 1e-16),
'prefix': ''
}
cf_x_fluct_pars = {
'target_subdomain': x_space,
'scale': (1e-2, 1e-16),
'cutoff': (1., 1e-16),
'loglogslope': (-2.0, 1e-16)
}
init_model(cf_make_pars, cf_x_fluct_pars, matern=True)
model domain keys: ('cutoff', 'loglogslope', 'scale', 'xi', 'zeromode')
# Show neutral field
eval_model(cf_make_pars, cf_x_fluct_pars, "Neutral Field", matern=True)
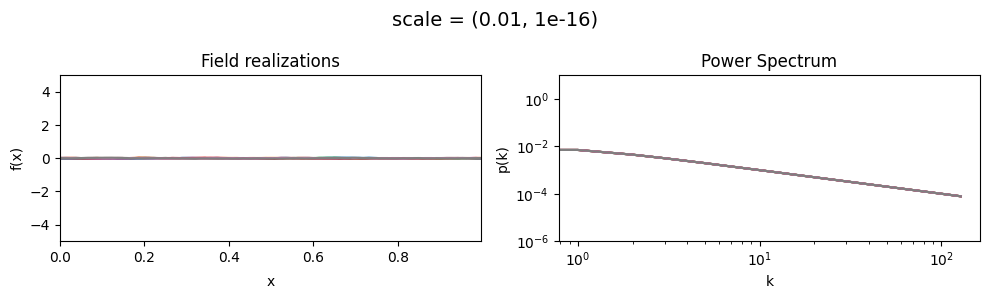
The scale parameters of add_fluctuations_matern()#
determine the overall amplitude scaling factor of fluctuations in the target subdomain
for which add_fluctuations_matern is called.
It does not set the absolute amplitude, which depends on all other parameters, too.
scale[0] set the average amplitude scaling factor of the fields’ fluctuations along the given dimension,
scale[1] sets the width and shape of the scaling factor distribution.
The scaling factor is modelled as being log-normally distributed,
see The Moment-Matched Log-Normal Distribution above for details.
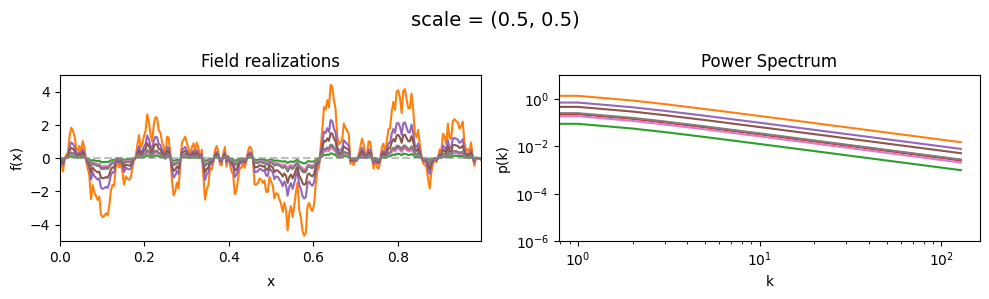
scale mean:#
vary_parameter('scale', [(0.01, 1e-16), (0.1, 1e-16), (1.0, 1e-16)], samples_vary_in='xi', matern=True)
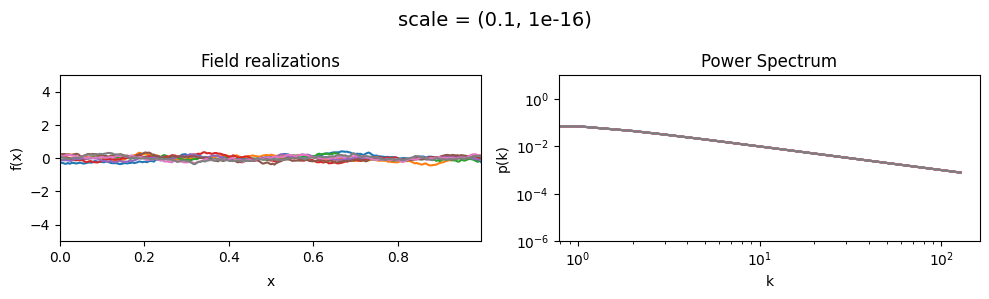
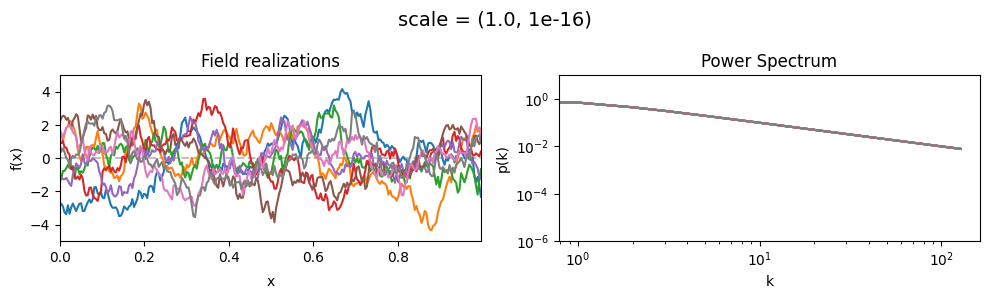
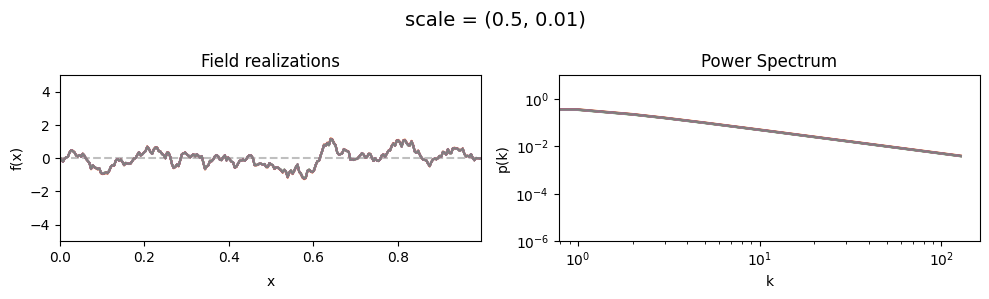
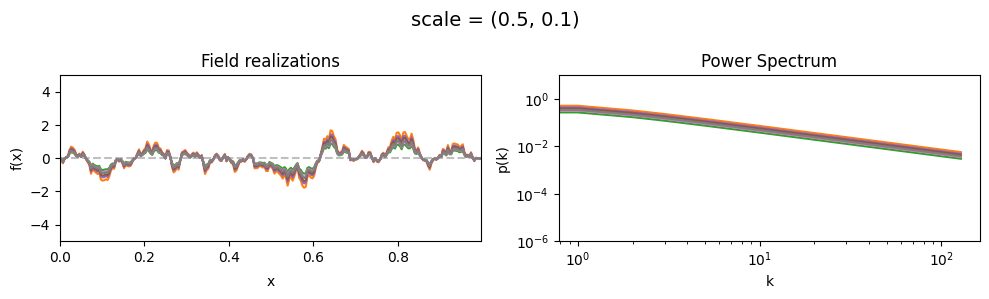
scale std:#
vary_parameter('scale', [(0.5, 0.01), (0.5, 0.1), (0.5, 0.5)], samples_vary_in='scale', matern=True)
cf_x_fluct_pars['scale'] = (0.5, 1e-16)
The loglogslope parameters of add_fluctuations_matern()#
determine the slope of the loglog-linear (power law) component of the power spectrum.
loglogslope[0] set the average power law exponent of the fields’ power spectrum along the given dimension,
loglogslope[1] sets the width and shape of the exponent distribution.
The loglogslope is modelled to be normally distributed.
loglogslope mean:#
vary_parameter('loglogslope', [(-4.0, 1e-16), (-2.0, 1e-16), (-1.0, 1e-16)], samples_vary_in='xi', matern=True)
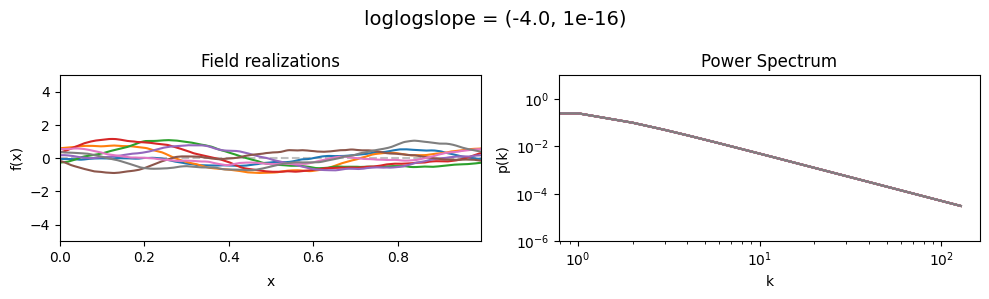
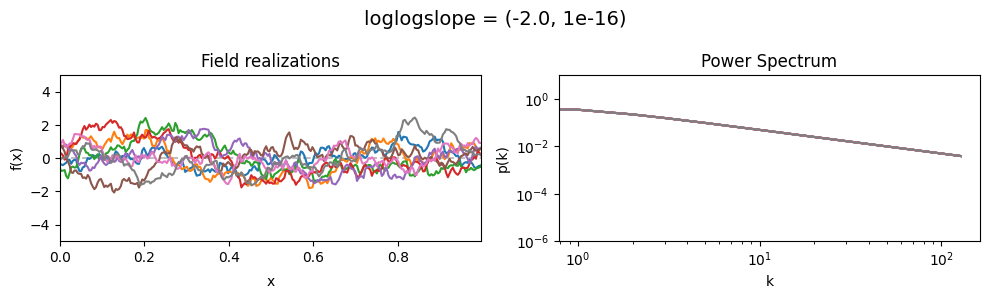
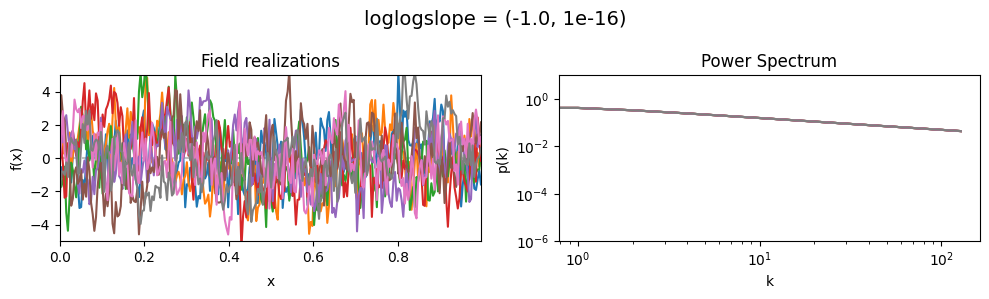
As one can see, the field amplitude also depends on the loglogslope parameter.
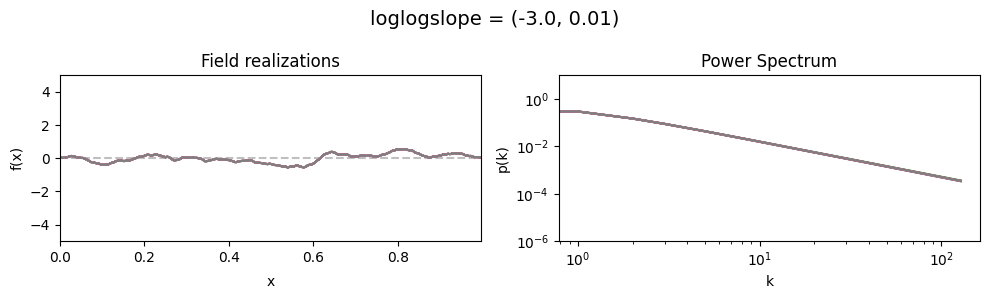
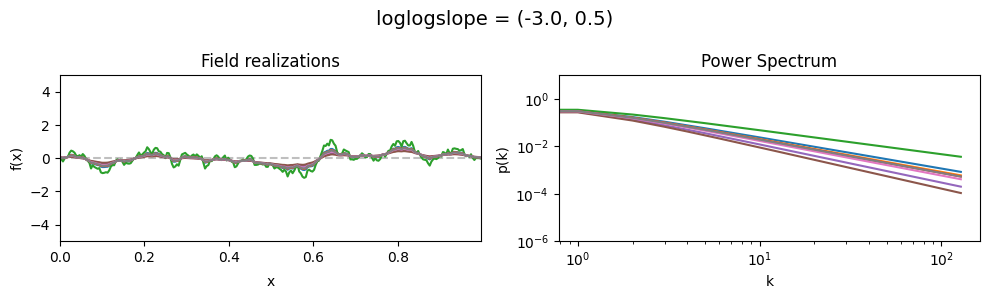
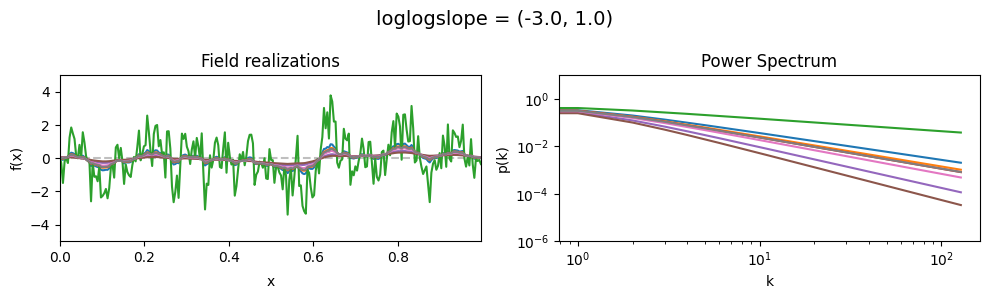
loglogslope std:#
vary_parameter('loglogslope', [(-3., 0.01), (-3., 0.5), (-3., 1.0)], samples_vary_in='loglogslope', matern=True)
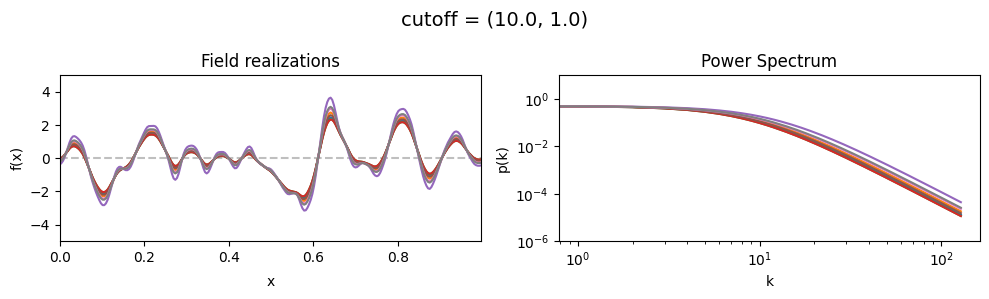
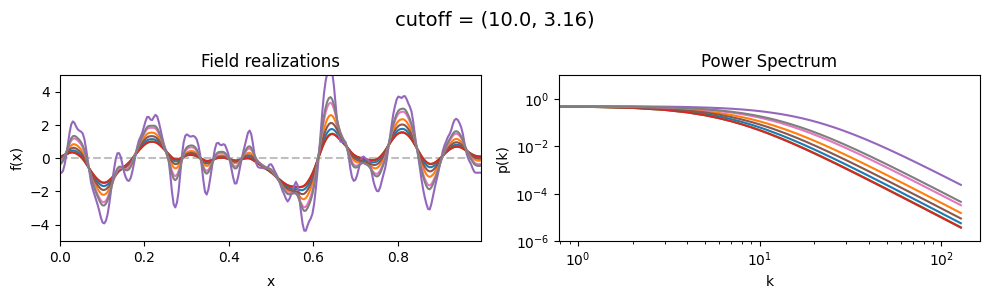
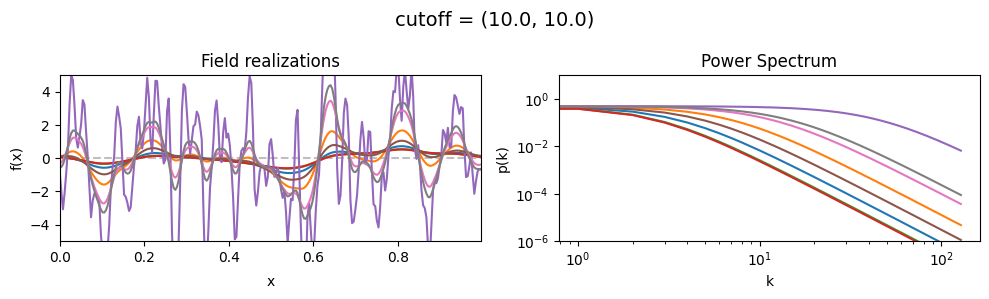
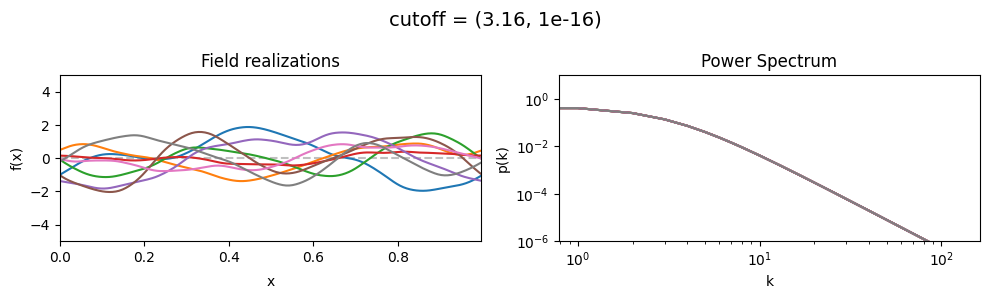
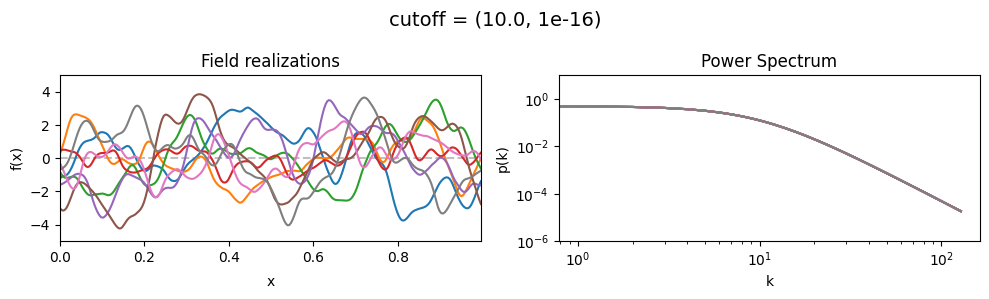
The cutoff parameters of add_fluctuations_matern()#
determines at what wavevector length the power spectrum should transition from constant power
to following the powerlaw set by loglogslope.
cutoff[0] set the average wavevector length at which the power spectrum transition occurs,
cutoff[1] sets the width and shape of the transition wavevector length distribution.
The cutoff is modelled as being log-normally distributed,
see The Moment-Matched Log-Normal Distribution above for details.
cutoff mean:#
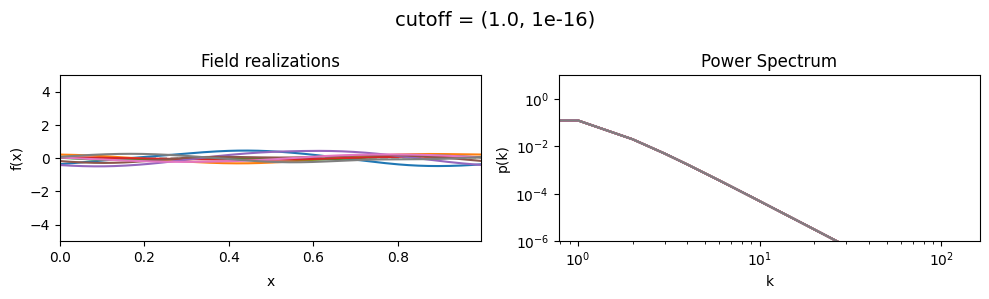
cf_x_fluct_pars['loglogslope'] = (-8.0, 1e-16)
vary_parameter('cutoff', [(1.0, 1e-16), (3.16, 1e-16), (10.0, 1e-16)], samples_vary_in='xi', matern=True)
cutoff std:#
vary_parameter('cutoff', [(10., 1.0), (10., 3.16), (10., 10.)], samples_vary_in='cutoff', matern=True)